FGFR2 fusions in iCCA
Genetic alterations of fibroblast growth factor receptor (FGFR) exist across different tumor types, including intrahepatic cholangiocarcinoma (iCCA)
FGFR has emerged as a tumorigenic driver in cancers such as iCCA, urothelial carcinoma, myeloid/lymphoid neoplasms, and other malignancies.1-3
Genetic alterations have been observed in all FGFR subtypes (FGFR1, FGFR2, FGFR3, and FGFR4). Alterations include point mutations, gene amplifications, and chromosomal rearrangements that may result in fusion proteins, including FGFR2 fusions.4

In iCCA, FGFR2 fusions have been identified as key oncogenic drivers in 10% to 16% of patients7-9
FGFR2 fusions are found almost exclusively in iCCA.5,8 When they occur, they cause constitutive FGFR2 signaling, which in turn contributes to a variety of tumorigenic processes.10,11

FGFR2 fusions are detectable early in disease progression, elevating the importance of identifying these alterations in patients at diagnosis.12
Along with FGFR2 fusions, other clinically significant alterations have been identified in iCCA
Clinically significant genomic alterations have been identified in ~50% of iCCA patients, enabling a more individualized picture of each patient's disease biology.5,9,13,14 The most common clinically significant genomic alterations include FGFR2 fusions and isocitrate dehydrogenase (IDH) mutations.9
Clinically significant genomic alterations are abundant in iCCA*
Tap an alteration to learn more
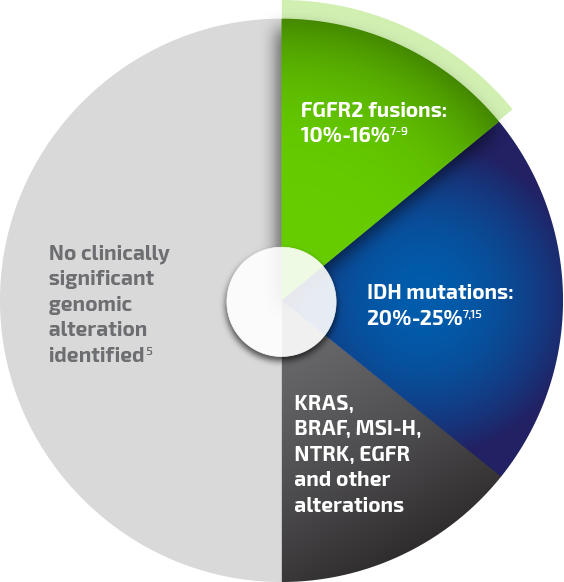
Percentages reported for individual alterations are from different primary analyses and are not all drawn from a single population of patients with iCCA.
*Although FGFR2 fusions and IDH mutations tend to be mutually exclusive in iCCA, mutations in the "other alterations" category can be found in tumors that also carry other clinically significant mutations or genomic alterations.
BRAF, v-raf murine sarcoma viral oncogene homolog B1; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene homolog; MSI-H, microsatellite instability-high; NTRK, neurotrophic tyrosine receptor kinase.
FGFR2 fusions:
Have been discovered to drive iCCA tumor growth through ligand-independent receptor activity, which leads to overactive signaling1,16,17
Are detectable early in disease progression12
IDH mutations:
Somatic mutations in metabolic enzymes that ultimately affect epigenetic regulation as well as cell differentiation and metabolism16,18
Other:
BRAF mutations: ~3%-7%19
Activating mutations in the signal transduction cascade that affect cell proliferation, secretion, and differentiation16,20
KRAS mutations: ~9%-24%19
Activating mutations of the proto-oncogene encoding for a protein belonging to the GTPase family16,20
MSI-H status: <1%-2.5%5
Although not a single genomic alteration, MSI-H (microsatellite instability-high) status reflects a propensity for mutation due to abnormalities of the DNA sequence. It may be identified by expression of genes such as MLH1, PMS2, MSH2, and MSH621,22
NTRK fusion: <1%-4%23
Chimeric NTRK fusion proteins promote tumorigenesis through constitutive ligand-free activation of intracellular biological pathways and signal transduction cascades that control cell-cycle progression, proliferation, apoptosis, and survival.24
EGFR mutations: ~1.5%-2%19
EGFR activation leads to downstream activation of mitogen-activated protein kinase, a well-known oncogenic signaling pathway.25
REFERENCES: 1. Babina IS, Turner NC. Nat Rev Cancer. 2017;17(5):318-332. 2. Pandith AA, Shah ZA, Siddiqi MA. Urol Oncol. 2013;31(4):398-406. 3. Gallo LH, Nelson KN, Meyer AN, Donoghue DJ. Cytokine Growth Factor Rev. 2015;26(4):425-449. 4. Jain A, Borad MJ, Kelley RK, et al. JCO Precis Oncol. 2018;2:1-12. 5. Lowery MA, Ptashkin R, Jordan E, et al. Clin Cancer Res. 2018;24(17):4154-4161. 6. Shibata T, Arai Y, Totoki Y. Cancer Sci. 2018;109(5):1282-1291. 7. Farshidfar F, Zheng S, Gingras MC, et al. Cell Rep. 2017;18(11):2780-2794. 8. Graham RP, Barr Fritcher EG, Pestova E, et al. Hum Pathol. 2014;45(8):1630-1638. 9. Ross JS, Wang K, Gay L, et al. Oncologist. 2014;19(3):235-242. 10. Arai Y, Totoki Y, Hosoda F, et al. Hepatology. 2014;59(4):1427-1434. 11. Borad MJ, Champion MD, Egan JB, et al. PLoS Genet. 2014;10(2):e1004135. 12. Rizvi S, Borad MJ. J Gastrointest Oncol. 2016;7(5):789-796. 13. Sia D, Losic B, Moeini A, et al. Nat Commun. 2015;6:6087. 14. Chun SY, Javle M. Cancer Contr. 2017;24(3):1-7. 15. Churi CR, Shroff R, Wang Y, et al. PLoS ONE. 2014;9(12):e115383. doi:10.1371/journal.pone.0115383. 16. Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Clin Cancer Res. 2015;22(2):291-300. 17. Touat M, Ileana E, Postel-Vinay S, André F, Soria JC. Clin Cancer Res. 2015;21(12):2684-2694. 18. Mondesir J, Willekens C, Touat M, de Botton S. J Blood Med. 2016;7:171-180. 19. Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. Cancer Discov. 2017;7(9):943-962. 20. Pellino A, Loupakis F, Cadamuro M, et al. Transl Gastroenterol Hepatol. 2018;3:40. doi:10.21037/tgh.2018.07.02. 21. Winkelmann R, Schneider M, Hartmann S, et al. Int J Mol Sci. 2018;19(5):1421. doi:10.3390/ijms19051421. 22. Momoi H, Itoh T, Nozaki Y, et al. J Hepatol. 2001;35(2):235-244. 23. Solomon JP, Linkov I, Rosado A, et al. Mod Pathol. 2019. [Epub ahead of print]. doi: 10.1038/s41379-019-0324-7. 24. Kheder ES, Hong DS. Clin Cancer Res. 2018;24(23):5807-5814. 25. Rizvi S, Borad MJ, Patel T, Gores GJ. Semin Liver Dis. 2014;34(4):456-464.
